Introduction
R Markdown is a great tool to use for creating reports, presentations and even websites that contain evaluated and rendered code. This can help us immensely when presenting data science type of work to audiences, while still being able to version control the content creation process.
One of the challenges that stay is reproducibility of the rendered results. In this post, I will list a few sources of reproducibility issues I came across and how I tried to solve them. As an introductory disclaimer, this post is not an exhaustive guide but merely a retrospect on the issues I faced and how I tackled them when writing posts for this blog using blogdown.
For this post, we would consider an R Markdown document reproducible if we can be sure that it produces identical rendered output as long as the content of the Rmd document, the data used within it and the rendering function stay the same.
This sounds like a reasonable thing to ask, however, there are many ways in which this assumption can be broken. And they are not always trivial - that is, unless your name is Yihui Xie :)
My guess is that you upgraded Pandoc first, saw the diffs, then updated the rmarkdown package (from a very old version), which now defaults the
— Yihui Xie (@xieyihui) January 30, 2019htmloutput format tohtml4(which generates<div>) instead of Pandoc's defaulthtml5(which generates<section>).
We will try to categorize some of the reasons into groups.
Contents
Output changes caused by code chunks not behaving reproducibly
The first group is the one that we have full control over, as it directly relates to the content of the code chunks in our R Markdown document.
Simple examples that showcase the issue
- Obviously, the output can change each time we run this chunk:
```{r}
Sys.time()
```- Another scenario is code chunks that make use of random number generation. If we render an Rmd that contains this code chunk multiple times we will get different results, unless we take precautions:
```{r}
runif(5)
```- Running timing (benchmarking) code is almost certain to produce different results each time it is run, even though the benchmarked code is identical:
```{r}
system.time(runif(1e6))
```Solution 1 - Remove output change source from the chunks
The most obvious and clean way to tackle the issue is to change our code such that the source of variability is removed. For random number generation, this can be achieved by setting a seed, e.g using set.seed(). This solution can get more complex as the scope increases - if you are interested in reading more on the topic of reproducibility with RNG, look here.
Solution 2 - Run the code once and store the results
For some code, such as benchmarking, fixing the code such that the output does not change is very difficult in principle, therefore we must find a workaround that would ensure the results stay untouched. One approach is to run the code once, store the results and do not run the code again on render. Some ways to do that:
Using the cache=TRUE chunk option
In practice, this can be done nicely by using the cache=TRUE option, which provides this behavior and also makes sure that the cache is updated automatically when the code chunk changes, so the correctness of results is ensured. Exceptions to this exist for some special cases, read the details in the knitr manual for a deeper understanding. Here is an example chunk using that option:
```{r cache=TRUE}
system.time(runif(1e6))
```Storing a needed representation of the object directly in the Rmd
One property of knitr’s caching that could be considered a downside is that the cache storage uses binary files, which while being a completely natural choice is not the best for version control. Especially when this is a concern and the code chunk outputs are small in size, other options may also be considered.
One such example would be to save a needed representation of the result directly in the Rmd and use eval=FALSE on the code chunk. This comes with trade-offs too, notably we must pay attention for the chunk changes, as there would be no automated update similar to the one provided by knitr’s cache mechanism.
As an example, we could rewrite the chunk above into two chunks like so - the first chunk shows the code in the output without running, the second makes sure that the results that we store gets shown (but the code does not):
```{r eval=FALSE}
system.time(runif(1e6))
```
```{r echo=FALSE}
# This is pre-calculated and just shown to keep the output static
structure(
c(0.081, 0.003, 0.084, 0, 0)
class = "proc_time"
.Names = c("user.self", "sys.self", "elapsed", "user.child", "sys.child")
)
```The content of the second chunk can be obtained by using dput(system.time(runif(1e6))). Naturally, this may become quite impractical to use with bigger objects.
Storing the rendered output directly in the Rmd
Another variation of the above would be to not even bother with obtaining the representation of the output via evaluating a code chunk, but just placing the rendered output itself into the Rmd directly. This comes with the same downsides as the previous approach with some extras. Mainly, we need to create a format-specific output, meaning this approach can be considered only if the output format is fixed and will not change.
For example, we can be reasonably sure that for a blogdown website the output will be HTML. In case we are ok with all those trade-offs, we can place the following into our .Rmd to represent the above code chunks in HTML:
<pre><code>
## user system elapsed
## 0.081 0.003 0.084
</code></pre>Output changes caused by different package versions
The tricky issues start when our Rmd content is actually reproducible under our current local setup, however the rendered output changes with a different setup, often with changed package versions. A concrete real-life example is when an update of the highcharter package slightly tweaks the output:
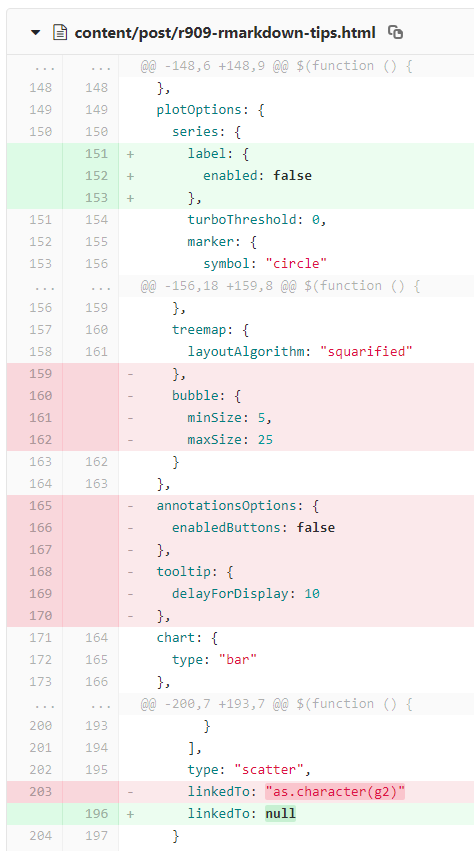
Slightly different highchart representation
Solution - Package version management, e.g. with packrat
Solving issues with package versions is a broad topic, so we will only mention one that is relatively easy to use, especially with RStudio - Packrat. Packrat is an R package that works by creating a separate library of packages on a project basis.
We can create separate R projects for Rmd files with shared package dependencies, or even a separate project per Rmd. This will ensure that we always have the intended set of packages loaded and used for them.
As all of the above, apart from the extra overhead of using it, this approach can have its caveats as well. Using packrat to manage a blogdown site or a bookdown book likely means that all of the site posts or book chapters will use a shared package library, which may not be granular enough for all use cases.
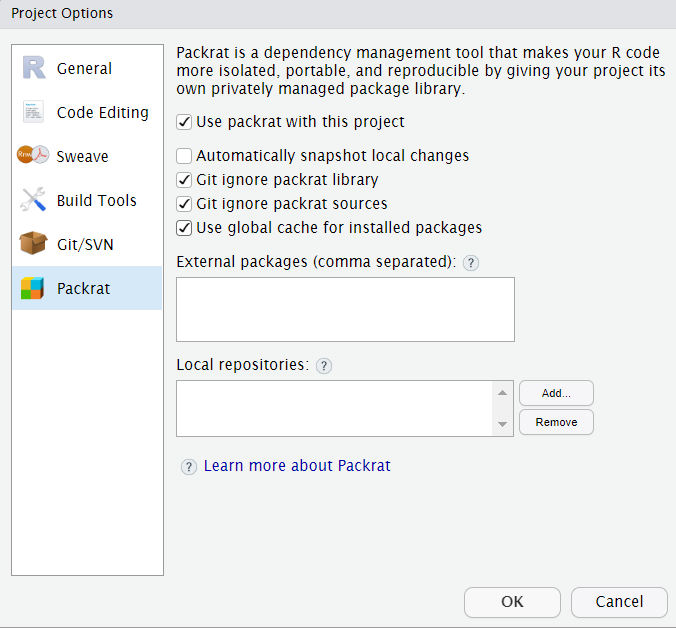
Using packrat with a project
Care also has to be taken to make sure that the packrat managed libraries are used when rendering the content, i.e. by ensuring that the packrat/init.R is sourced before the rendering happens.
Output changes caused by changed system dependencies
Going into deeper circles of the dreaded dependency hell, even if we manage our R packages with care, the system dependencies can still cause behavior that would change our output in an unintended way. In the case of R Markdown and knitr the most notable dependency of this type is Pandoc, the powerhouse behind R Markdown.
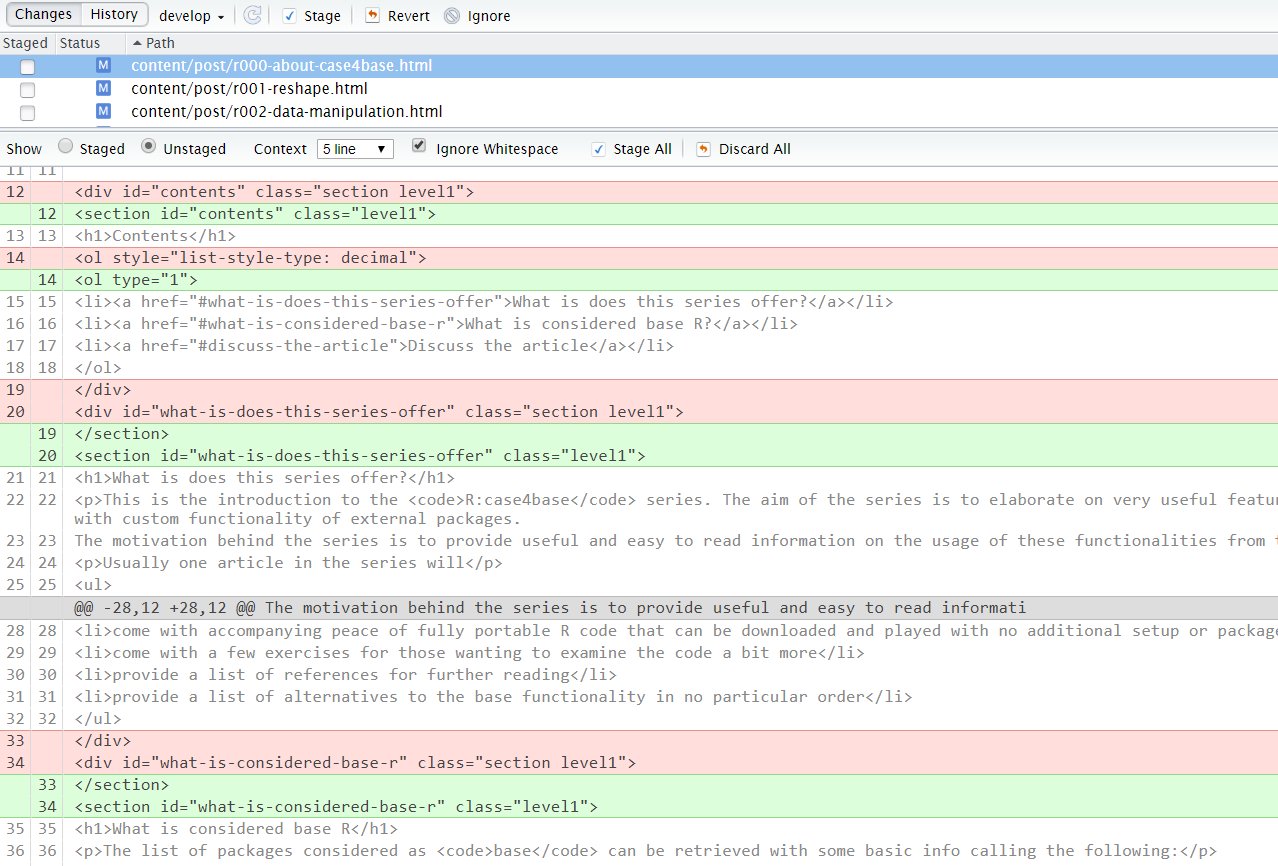
Why would this happen?
A way this dependency can change is for example when updating RStudio Server, which comes bundled with a certain version of Pandoc:
- RStudio Server v1.1.453 comes with Pandoc 1.19.2.1
- RStudio Server v1.2.1234 comes with Pandoc 2.3.1
- You may have an even newer version on Pandoc installed if you got it separately. As of the date of writing this, the latest stable release of Pandoc is 2.5
Solution - To containerization and beyond
Solving system dependencies is a tricky task made easier by a few tools but it is way out of the scope of this post, so we will only briefly list some options:
- Containerize your environment with an implementation of Operating-system level virtualization such as the ever so popular Docker
- Create a predefined VM setup, for example using configuration management software such as Ansible or Puppet